Deficiency for the ubiquitin ligase UBE3B in a blepharophimosis-ptosis-intellectual-disability syndrome
- PMID: 23200864
- PMCID: PMC3516591
- DOI: 10.1016/j.ajhg.2012.10.011
Deficiency for the ubiquitin ligase UBE3B in a blepharophimosis-ptosis-intellectual-disability syndrome
Abstract
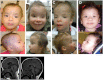
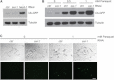
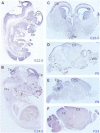
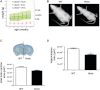
Ubiquitination plays a crucial role in neurodevelopment as exemplified by Angelman syndrome, which is caused by genetic alterations of the ubiquitin ligase-encoding UBE3A gene. Although the function of UBE3A has been widely studied, little is known about its paralog UBE3B. By using exome and capillary sequencing, we here identify biallelic UBE3B mutations in four patients from three unrelated families presenting an autosomal-recessive blepharophimosis-ptosis-intellectual-disability syndrome characterized by developmental delay, growth retardation with a small head circumference, facial dysmorphisms, and low cholesterol levels. UBE3B encodes an uncharacterized E3 ubiquitin ligase. The identified UBE3B variants include one frameshift and two splice-site mutations as well as a missense substitution affecting the highly conserved HECT domain. Disruption of mouse Ube3b leads to reduced viability and recapitulates key aspects of the human disorder, such as reduced weight and brain size and a downregulation of cholesterol synthesis. We establish that the probable Caenorhabditis elegans ortholog of UBE3B, oxi-1, functions in the ubiquitin/proteasome system in vivo and is especially required under oxidative stress conditions. Our data reveal the pleiotropic effects of UBE3B deficiency and reinforce the physiological importance of ubiquitination in neuronal development and function in mammals.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Expanding the clinical and mutational spectrum of Kaufman oculocerebrofacial syndrome with biallelic UBE3B mutations.Hum Genet. 2014 Jul;133(7):939-49. doi: 10.1007/s00439-014-1436-2. Epub 2014 Mar 11. Hum Genet. 2014. PMID: 24615390
-
Loss of function of the E3 ubiquitin-protein ligase UBE3B causes Kaufman oculocerebrofacial syndrome.J Med Genet. 2013 Aug;50(8):493-9. doi: 10.1136/jmedgenet-2012-101405. Epub 2013 May 17. J Med Genet. 2013. PMID: 23687348 Free PMC article.
-
Blepharophimosis-ptosis-intellectual disability syndrome: A report of nine Egyptian patients with further expansion of phenotypic and mutational spectrum.Am J Med Genet A. 2020 Dec;182(12):2857-2866. doi: 10.1002/ajmg.a.61857. Epub 2020 Sep 19. Am J Med Genet A. 2020. PMID: 32949109
-
Further phenotypic characterization of Kaufman oculocerebrofacial syndrome: report of five new cases and literature review.Clin Dysmorphol. 2019 Oct;28(4):175-183. doi: 10.1097/MCD.0000000000000282. Clin Dysmorphol. 2019. PMID: 31162149 Review.
-
Blepharophimosis-ptosis-epicanthus inversus syndrome (BPES).Acta Ophthalmol Scand. 1996 Feb;74(1):45-7. doi: 10.1111/j.1600-0420.1996.tb00680.x. Acta Ophthalmol Scand. 1996. PMID: 8689480 Review.
Cited by
-
Loss of caspase-2 augments lymphomagenesis and enhances genomic instability in Atm-deficient mice.Proc Natl Acad Sci U S A. 2013 Dec 3;110(49):19920-5. doi: 10.1073/pnas.1311947110. Epub 2013 Nov 18. Proc Natl Acad Sci U S A. 2013. PMID: 24248351 Free PMC article.
-
Kaufman oculo-cerebro-facial syndrome in a child with small and absent terminal phalanges and absent nails.J Hum Genet. 2017 Apr;62(4):465-471. doi: 10.1038/jhg.2016.151. Epub 2016 Dec 22. J Hum Genet. 2017. PMID: 28003643 Free PMC article.
-
Early Origin and Evolution of the Angelman Syndrome Ubiquitin Ligase Gene Ube3a.Front Cell Neurosci. 2017 Mar 7;11:62. doi: 10.3389/fncel.2017.00062. eCollection 2017. Front Cell Neurosci. 2017. PMID: 28326016 Free PMC article. Review.
-
Accurate phenotypic classification and exome sequencing allow identification of novel genes and variants associated with adult-onset hearing loss.PLoS Genet. 2023 Nov 27;19(11):e1011058. doi: 10.1371/journal.pgen.1011058. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 38011198 Free PMC article.
-
RNF13 Dileucine Motif Variants L311S and L312P Interfere with Endosomal Localization and AP-3 Complex Association.Cells. 2021 Nov 6;10(11):3063. doi: 10.3390/cells10113063. Cells. 2021. PMID: 34831286 Free PMC article.
References
-
- Kawabe H., Brose N. The role of ubiquitylation in nerve cell development. Nat. Rev. Neurosci. 2011;12:251–268. - PubMed
-
- Tai H.C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008;9:826–838. - PubMed
-
- Komander D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009;37:937–953. - PubMed
-
- Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A., Chanda S.K., Batalov S., Joazeiro C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

