Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability
- PMID: 21888914
- PMCID: PMC3321355
- DOI: 10.1016/j.jmb.2011.08.029
Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability
Abstract
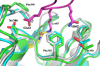
AMSH plays a critical role in the ESCRT (endosomal sorting complexes required for transport) machinery, which facilitates the down-regulation and degradation of cell-surface receptors. It displays a high level of specificity toward cleavage of Lys63-linked polyubiquitin chains, the structural basis of which has been understood recently through the crystal structure of a highly related, but ESCRT-independent, protein AMSH-LP (AMSH-like protein). We have determined the X-ray structure of two constructs representing the catalytic domain of AMSH: AMSH244, the JAMM (JAB1/MPN/MOV34)-domain-containing polypeptide segment from residues 244 to 424, and AMSH219(E280A), an active-site mutant, Glu280 to Ala, of the segment from 219 to 424. In addition to confirming the expected zinc coordination in the protein, the structures reveal that the catalytic domains of AMSH and AMSH-LP are nearly identical; however, guanidine-hydrochloride-induced unfolding studies show that the catalytic domain of AMSH is thermodynamically less stable than that of AMSH-LP, indicating that the former is perhaps structurally more plastic. Much to our surprise, in the AMSH219(E280A) structure, the catalytic zinc was still held in place, by the compensatory effect of an aspartate from a nearby loop moving into a position where it could coordinate with the zinc, once again suggesting the plasticity of AMSH. Additionally, a model of AMSH244 bound to Lys63-linked diubiquitin reveals a type of interface for the distal ubiquitin significantly different from that seen in AMSH-LP. Altogether, we believe that our data provide important insight into the structural difference between the two proteins that may translate into the difference in their biological function.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



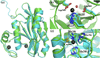
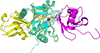



Similar articles
-
Mechanism of recruitment and activation of the endosome-associated deubiquitinase AMSH.Biochemistry. 2013 Nov 5;52(44):7818-29. doi: 10.1021/bi401106b. Epub 2013 Oct 23. Biochemistry. 2013. PMID: 24151880 Free PMC article.
-
NMR Reveals the Interplay among the AMSH SH3 Binding Motif, STAM2, and Lys63-Linked Diubiquitin.J Mol Biol. 2016 Nov 6;428(22):4544-4558. doi: 10.1016/j.jmb.2016.10.002. Epub 2016 Oct 8. J Mol Biol. 2016. PMID: 27725184
-
Crystal structure of the Thr316Ala mutant of a yeast JAMM deubiquitinase: implication of active-site loop dynamics in catalysis.Acta Crystallogr F Struct Biol Commun. 2021 Jun 1;77(Pt 6):163-170. doi: 10.1107/S2053230X21005124. Epub 2021 May 24. Acta Crystallogr F Struct Biol Commun. 2021. PMID: 34100774 Free PMC article.
-
Structure and function of MPN (Mpr1/Pad1 N-terminal) domain-containing proteins.Curr Protein Pept Sci. 2014;15(5):504-17. doi: 10.2174/1389203715666140221095109. Curr Protein Pept Sci. 2014. PMID: 24555901 Review.
-
Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination.Cell Biochem Biophys. 2011 Jun;60(1-2):39-46. doi: 10.1007/s12013-011-9181-9. Cell Biochem Biophys. 2011. PMID: 21448666 Review.
Cited by
-
Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4.Nat Struct Mol Biol. 2018 Jul;25(7):616-622. doi: 10.1038/s41594-018-0085-x. Epub 2018 Jul 2. Nat Struct Mol Biol. 2018. PMID: 29967539 Free PMC article.
-
Dynamics of an Active-Site Flap Contributes to Catalysis in a JAMM Family Metallo Deubiquitinase.Biochemistry. 2015 Oct 6;54(39):6038-51. doi: 10.1021/acs.biochem.5b00631. Biochemistry. 2015. PMID: 26368668 Free PMC article.
-
Validation and correction of Zn-CysxHisy complexes.Acta Crystallogr D Struct Biol. 2016 Oct 1;72(Pt 10):1110-1118. doi: 10.1107/S2059798316013036. Epub 2016 Sep 15. Acta Crystallogr D Struct Biol. 2016. PMID: 27710932 Free PMC article.
-
Ubiquitin and its relatives as wizards of the endolysosomal system.J Cell Sci. 2023 Feb 15;136(4):jcs260101. doi: 10.1242/jcs.260101. Epub 2023 Feb 24. J Cell Sci. 2023. PMID: 36825571 Free PMC article.
-
Structure and Function of the 26S Proteasome.Annu Rev Biochem. 2018 Jun 20;87:697-724. doi: 10.1146/annurev-biochem-062917-011931. Epub 2018 Apr 13. Annu Rev Biochem. 2018. PMID: 29652515 Free PMC article. Review.
References
-
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. - PubMed
-
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. - PubMed
-
- Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem. 2010;54:69–87. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

