Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss
- PMID: 21532572
- PMCID: PMC3102765
- DOI: 10.1038/ng.830
Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss
Abstract
DNA methyltransferase 1 (DNMT1) is crucial for maintenance of methylation, gene regulation and chromatin stability. DNA mismatch repair, cell cycle regulation in post-mitotic neurons and neurogenesis are influenced by DNA methylation. Here we show that mutations in DNMT1 cause both central and peripheral neurodegeneration in one form of hereditary sensory and autonomic neuropathy with dementia and hearing loss. Exome sequencing led to the identification of DNMT1 mutation c.1484A>G (p.Tyr495Cys) in two American kindreds and one Japanese kindred and a triple nucleotide change, c.1470-1472TCC>ATA (p.Asp490Glu-Pro491Tyr), in one European kindred. All mutations are within the targeting-sequence domain of DNMT1. These mutations cause premature degradation of mutant proteins, reduced methyltransferase activity and impaired heterochromatin binding during the G2 cell cycle phase leading to global hypomethylation and site-specific hypermethylation. Our study shows that DNMT1 mutations cause the aberrant methylation implicated in complex pathogenesis. The discovered DNMT1 mutations provide a new framework for the study of neurodegenerative diseases.
Figures







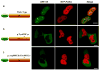
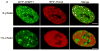


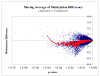
Comment in
-
DNMT1 links aberrant DNA methylation to hereditary sensory neuropathy.Clin Genet. 2011 Sep;80(3):240-1. doi: 10.1111/j.1399-0004.2011.01752.x. Clin Genet. 2011. PMID: 21762444 No abstract available.
Similar articles
-
Aberrant signature methylome by DNMT1 hot spot mutation in hereditary sensory and autonomic neuropathy 1E.Epigenetics. 2014 Aug;9(8):1184-93. doi: 10.4161/epi.29676. Epub 2014 Jul 7. Epigenetics. 2014. PMID: 25033457 Free PMC article.
-
DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation.Hum Mol Genet. 2017 Apr 15;26(8):1522-1534. doi: 10.1093/hmg/ddx057. Hum Mol Genet. 2017. PMID: 28334952 Free PMC article.
-
Defects of mutant DNMT1 are linked to a spectrum of neurological disorders.Brain. 2015 Apr;138(Pt 4):845-61. doi: 10.1093/brain/awv010. Epub 2015 Feb 11. Brain. 2015. PMID: 25678562 Free PMC article.
-
Effect of Disease-Associated Germline Mutations on Structure Function Relationship of DNA Methyltransferases.Genes (Basel). 2019 May 14;10(5):369. doi: 10.3390/genes10050369. Genes (Basel). 2019. PMID: 31091831 Free PMC article. Review.
-
An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function.Epigenetics. 2012 Sep;7(9):994-1007. doi: 10.4161/epi.21568. Epub 2012 Aug 16. Epigenetics. 2012. PMID: 22894906 Free PMC article. Review.
Cited by
-
Modifiers and Readers of DNA Modifications and Their Impact on Genome Structure, Expression, and Stability in Disease.Front Genet. 2016 Jun 21;7:115. doi: 10.3389/fgene.2016.00115. eCollection 2016. Front Genet. 2016. PMID: 27446199 Free PMC article. Review.
-
Analysis of DNMT1 gene variants in progression of neural tube defects-an in silico to in vitro approach.Biosci Rep. 2022 Dec 22;42(12):BSR20220998. doi: 10.1042/BSR20220998. Biosci Rep. 2022. PMID: 36394275 Free PMC article.
-
DNA methylation and its basic function.Neuropsychopharmacology. 2013 Jan;38(1):23-38. doi: 10.1038/npp.2012.112. Epub 2012 Jul 11. Neuropsychopharmacology. 2013. PMID: 22781841 Free PMC article. Review.
-
Dynamics of DNA methylation in aging and Alzheimer's disease.DNA Cell Biol. 2012 Oct;31 Suppl 1(Suppl 1):S42-8. doi: 10.1089/dna.2011.1565. Epub 2012 Feb 7. DNA Cell Biol. 2012. PMID: 22313030 Free PMC article. Review.
-
Dnmt1-dependent DNA methylation is essential for photoreceptor terminal differentiation and retinal neuron survival.Cell Death Dis. 2012 Nov 22;3(11):e427. doi: 10.1038/cddis.2012.165. Cell Death Dis. 2012. PMID: 23171847 Free PMC article.
References
-
- Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. - PubMed
-
- Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–9. - PubMed
-
- Tohgi H, et al. Reduction with age in methylcytosine in the promoter region -224 approximately -101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999;70:288–92. - PubMed
-
- Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–78. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- NS21328/NS/NINDS NIH HHS/United States
- K08 NS065007/NS/NINDS NIH HHS/United States
- R01 NS036797/NS/NINDS NIH HHS/United States
- P30DK084567/DK/NIDDK NIH HHS/United States
- R01 CA132878-04/CA/NCI NIH HHS/United States
- P30 DK084567/DK/NIDDK NIH HHS/United States
- R01 NS021328/NS/NINDS NIH HHS/United States
- NS070298/NS/NINDS NIH HHS/United States
- UL1 RR024150/RR/NCRR NIH HHS/United States
- R01 CA132878/CA/NCI NIH HHS/United States
- R01 NS070298/NS/NINDS NIH HHS/United States
- R01 NS36797/NS/NINDS NIH HHS/United States
- K08 NS065007-02/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

