Structure of ATP-bound human ATP:cobalamin adenosyltransferase
- PMID: 17176040
- PMCID: PMC2532598
- DOI: 10.1021/bi061396f
Structure of ATP-bound human ATP:cobalamin adenosyltransferase
Abstract
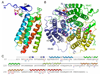
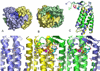
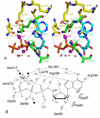
Mutations in the gene encoding human ATP:cobalamin adenosyltransferase (hATR) can result in the metabolic disorder known as methylmalonic aciduria (MMA). This enzyme catalyzes the final step in the conversion of cyanocobalamin (vitamin B12) to the essential human cofactor adenosylcobalamin. Here we present the 2.5 A crystal structure of ATP bound to hATR refined to an Rfree value of 25.2%. The enzyme forms a tightly associated trimer, where the monomer comprises a five-helix bundle and the active sites lie on the subunit interfaces. Only two of the three active sites within the trimer contain the bound ATP substrate, thereby providing examples of apo- and substrate-bound-active sites within the same crystal structure. Comparison of the empty and occupied sites indicates that twenty residues at the enzyme's N-terminus become ordered upon binding of ATP to form a novel ATP-binding site and an extended cleft that likely binds cobalamin. The structure explains the role of 20 invariant residues; six are involved in ATP binding, including Arg190, which hydrogen bonds to ATP atoms on both sides of the scissile bond. Ten of the hydrogen bonds are required for structural stability, and four are in positions to interact with cobalamin. The structure also reveals how the point mutations that cause MMA are deficient in these functions.
Figures
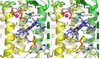
Similar articles
-
Ligand-binding by catalytically inactive mutants of the cblB complementation group defective in human ATP:cob(I)alamin adenosyltransferase.Mol Genet Metab. 2009 Nov;98(3):278-84. doi: 10.1016/j.ymgme.2009.06.014. Epub 2009 Jun 27. Mol Genet Metab. 2009. PMID: 19625202
-
The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.J Biol Chem. 2004 May 28;279(22):23646-53. doi: 10.1074/jbc.M401395200. Epub 2004 Mar 25. J Biol Chem. 2004. PMID: 15044458
-
Crystal structure of PduO-Type ATP:Cob(I)alamin adenosyltransferase from Bacillus cereus in a complex with ATP.Biochem Biophys Res Commun. 2011 May 13;408(3):417-21. doi: 10.1016/j.bbrc.2011.04.036. Epub 2011 Apr 13. Biochem Biophys Res Commun. 2011. PMID: 21514283
-
Adenosyltransferase: an enzyme and an escort for coenzyme B12?Trends Biochem Sci. 2005 Jun;30(6):304-8. doi: 10.1016/j.tibs.2005.04.008. Trends Biochem Sci. 2005. PMID: 15950874 Review.
-
Vitamin B12 in health and disease: part I--inherited disorders of function, absorption, and transport.Gastroenterologist. 1995 Dec;3(4):329-44. Gastroenterologist. 1995. PMID: 8775094 Review.
Cited by
-
Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12 as a Cofactor Conservation Strategy.J Am Chem Soc. 2018 Oct 17;140(41):13205-13208. doi: 10.1021/jacs.8b08659. Epub 2018 Oct 8. J Am Chem Soc. 2018. PMID: 30282455 Free PMC article.
-
Restricted role for methionine synthase reductase defined by subcellular localization.Mol Genet Metab. 2008 May;94(1):68-77. doi: 10.1016/j.ymgme.2007.11.019. Epub 2008 Jan 24. Mol Genet Metab. 2008. PMID: 18221906 Free PMC article.
-
Dissecting the role of critical residues and substrate preference of a Fatty Acyl-CoA Synthetase (FadD13) of Mycobacterium tuberculosis.PLoS One. 2009 Dec 21;4(12):e8387. doi: 10.1371/journal.pone.0008387. PLoS One. 2009. PMID: 20027301 Free PMC article.
-
Crystallization and preliminary X-ray crystallographic studies of a PduO-type ATP:cob(I)alamin adenosyltransferase from Bacillus cereus.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Jul 1;64(Pt 7):648-50. doi: 10.1107/S1744309108016552. Epub 2008 Jun 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18607099 Free PMC article.
-
FunSAV: predicting the functional effect of single amino acid variants using a two-stage random forest model.PLoS One. 2012;7(8):e43847. doi: 10.1371/journal.pone.0043847. Epub 2012 Aug 24. PLoS One. 2012. PMID: 22937107 Free PMC article.
References
-
- Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley and Sons; 1999.
-
- Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. - PubMed
-
- Kräutler B. B12 Coenzymes, the Central Theme. In: Kräutler B, Arigoni D, Golding BT, editors. Vitamin B12 and B12-Proteins. New York: Wiley-VCH; 1997. pp. 3–43.
-
- Drummond JT, Matthews RG. Cobalamin-dependent and cobalamin-independent methionine synthases in Escherichia coli: two solutions to the same chemical problem. Adv Exp Med Biol. 1993;338:687–692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

