The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP
- PMID: 16723661
- PMCID: PMC1484429
- DOI: 10.1261/rna.55406
The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP
Abstract
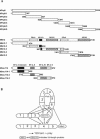
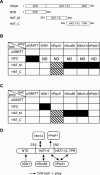
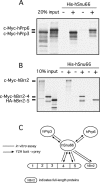
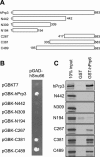
The human 25S U4/U6.U5 tri-snRNP is a major building block of the U2-type spliceosome and contains, in addition to the U4, U6, and U5 snRNAs, at least 30 distinct proteins. To learn more about the molecular architecture of the tri-snRNP, we have investigated interactions between tri-snRNP proteins using the yeast two-hybrid assay and in vitro binding assays, and, in addition, have identified distinct protein domains that are critical for the connectivity of this protein network in the human tri-snRNP. These studies revealed multiple interactions between distinct domains of the U5 proteins hPrp8, hBrr2 (a DExH/D-box helicase), and hSnu114 (a putative GTPase), which are key players in the catalytic activation of the spliceosome, during which the U4/U6 base-pairing interaction is disrupted and U4 is released from the spliceosome. Both the U5-specific, TPR/HAT-repeat-containing hPrp6 protein and the tri-snRNP-specific hSnu66 protein interact with several U5- and U4/U6-associated proteins, including hBrr2 and hPrp3, which contacts the U6 snRNA. Thus, both proteins are located at the interface between U5 and U4/U6 in the tri-snRNP complex, and likely play an important role in transmitting the activity of hBrr2 and hSnu114 in the U5 snRNP to the U4/U6 duplex during spliceosome activation. A more detailed analysis of these protein interactions revealed that different HAT repeats mediate interactions with specific hPrp6 partners. Taken together, data presented here provide a detailed picture of the network of protein interactions within the human tri-snRNP.
Figures



Similar articles
-
The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation.RNA. 2005 May;11(5):598-608. doi: 10.1261/rna.2300805. RNA. 2005. PMID: 15840814 Free PMC article.
-
Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing.EMBO J. 2002 Mar 1;21(5):1148-57. doi: 10.1093/emboj/21.5.1148. EMBO J. 2002. PMID: 11867543 Free PMC article.
-
The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase.RNA. 1997 Apr;3(4):344-55. RNA. 1997. PMID: 9085842 Free PMC article.
-
CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast.Curr Opin Struct Biol. 2016 Feb;36:48-57. doi: 10.1016/j.sbi.2015.12.005. Epub 2016 Jan 21. Curr Opin Struct Biol. 2016. PMID: 26803803 Free PMC article. Review.
-
The spliceosome.Bioessays. 1993 Sep;15(9):595-603. doi: 10.1002/bies.950150905. Bioessays. 1993. PMID: 8240312 Review.
Cited by
-
Proteomic analysis of cellular soluble proteins from human bronchial smooth muscle cells by combining nondenaturing micro 2DE and quantitative LC-MS/MS. 2. Similarity search between protein maps for the analysis of protein complexes.Electrophoresis. 2015 Sep;36(17):1991-2001. doi: 10.1002/elps.201400574. Epub 2015 Jul 20. Electrophoresis. 2015. PMID: 26031785 Free PMC article.
-
Spliceosome SNRNP200 Promotes Viral RNA Sensing and IRF3 Activation of Antiviral Response.PLoS Pathog. 2016 Jul 25;12(7):e1005772. doi: 10.1371/journal.ppat.1005772. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27454487 Free PMC article.
-
FgPrp4 Kinase Is Important for Spliceosome B-Complex Activation and Splicing Efficiency in Fusarium graminearum.PLoS Genet. 2016 Apr 8;12(4):e1005973. doi: 10.1371/journal.pgen.1005973. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27058959 Free PMC article.
-
Knocking Down Snrnp200 Initiates Demorphogenesis of Rod Photoreceptors in Zebrafish.J Ophthalmol. 2015;2015:816329. doi: 10.1155/2015/816329. Epub 2015 Jun 2. J Ophthalmol. 2015. PMID: 26137319 Free PMC article.
-
Mutations in spliceosomal proteins and retina degeneration.RNA Biol. 2017 May 4;14(5):544-552. doi: 10.1080/15476286.2016.1191735. Epub 2016 Jun 14. RNA Biol. 2017. PMID: 27302685 Free PMC article. Review.
References
-
- Achsel T., Ahrens K., Brahms H., Teigelkamp S., Lührmann R. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 1998;18:6756–6766. - PMC - PubMed
-
- Bartels C., Urlaub H., Lührmann R., Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 2003;278:28324–28334. - PubMed
-
- Blatch G.L., Lassle M. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
