Structural insight into the ligand specificity of a thermostable family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum
- PMID: 16336192
- PMCID: PMC1409695
- DOI: 10.1042/BJ20051780
Structural insight into the ligand specificity of a thermostable family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum
Abstract
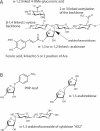
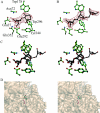
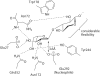
The digestion of the plant cell wall requires the concerted action of a diverse repertoire of enzyme activities. An important component of these hydrolase consortia are arabinofuranosidases, which release L-arabinofuranose moieties from a range of plant structural polysaccharides. The anaerobic bacterium Clostridium thermocellum, a highly efficient plant cell wall degrader, possesses a single alpha-L-arabinofuranosidase (EC 3.2.1.55), CtAraf51A, located in GH51 (glycoside hydrolase family 51). The crystal structure of the enzyme has been solved in native form and in 'Michaelis' complexes with both alpha-1,5-linked arabinotriose and alpha-1,3 arabinoxylobiose, both forming a hexamer in the asymmetric unit. Kinetic studies reveal that CtAraf51A, in contrast with well-characterized GH51 enzymes including the Cellvibrio japonicus enzyme [Beylot, McKie, Voragen, Doeswijk-Voragen and Gilbert (2001) Biochem. J. 358, 607-614], catalyses the hydrolysis of alpha-1,5-linked arabino-oligosaccharides and the alpha-1,3 arabinosyl side chain decorations of xylan with equal efficiency. The paucity of direct hydrogen bonds with the aglycone moiety and the flexible conformation adopted by Trp(178), which stacks against the sugar at the +1 subsite, provide a structural explanation for the plasticity in substrate specificity displayed by the clostridial arabinofuranosidase.
Figures
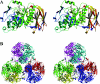
Similar articles
-
Molecular determinants of substrate specificity revealed by the structure of Clostridium thermocellum arabinofuranosidase 43A from glycosyl hydrolase family 43 subfamily 16.Acta Crystallogr D Struct Biol. 2016 Dec 1;72(Pt 12):1281-1289. doi: 10.1107/S205979831601737X. Epub 2016 Nov 29. Acta Crystallogr D Struct Biol. 2016. PMID: 27917828
-
Conservation in the mechanism of glucuronoxylan hydrolysis revealed by the structure of glucuronoxylan xylanohydrolase (CtXyn30A) from Clostridium thermocellum.Acta Crystallogr D Struct Biol. 2016 Nov 1;72(Pt 11):1162-1173. doi: 10.1107/S2059798316014376. Epub 2016 Oct 28. Acta Crystallogr D Struct Biol. 2016. PMID: 27841749
-
Heterologous expression and characterization of a putative glycoside hydrolase family 43 arabinofuranosidase from Clostridium thermocellum B8.Enzyme Microb Technol. 2018 Feb;109:74-83. doi: 10.1016/j.enzmictec.2017.09.014. Epub 2017 Oct 12. Enzyme Microb Technol. 2018. PMID: 29224629
-
GH62 arabinofuranosidases: Structure, function and applications.Biotechnol Adv. 2017 Nov 1;35(6):792-804. doi: 10.1016/j.biotechadv.2017.06.005. Epub 2017 Jun 29. Biotechnol Adv. 2017. PMID: 28669588 Review.
-
Glycosidase mechanisms.Curr Opin Chem Biol. 2002 Oct;6(5):619-29. doi: 10.1016/s1367-5931(02)00380-0. Curr Opin Chem Biol. 2002. PMID: 12413546 Review.
Cited by
-
Two Key Amino Acids Variant of α-l-arabinofuranosidase from Bacillus subtilis Str. 168 with Altered Activity for Selective Conversion Ginsenoside Rc to Rd.Molecules. 2021 Mar 19;26(6):1733. doi: 10.3390/molecules26061733. Molecules. 2021. PMID: 33808840 Free PMC article.
-
Biochemical and Molecular Dynamics Study of a Novel GH 43 α-l-Arabinofuranosidase/β-Xylosidase From Caldicellulosiruptor saccharolyticus DSM8903.Front Bioeng Biotechnol. 2022 Feb 11;10:810542. doi: 10.3389/fbioe.2022.810542. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35223784 Free PMC article.
-
A simple recipe for the non-expert bioinformaticist for building experimentally-testable hypotheses for proteins with no known homologs.J Struct Funct Genomics. 2012 Dec;13(4):185-200. doi: 10.1007/s10969-012-9141-7. Epub 2012 Sep 7. J Struct Funct Genomics. 2012. PMID: 22956349 Review.
-
A novel α-L-arabinofuranosidase of family 43 glycoside hydrolase (Ct43Araf) from Clostridium thermocellum.PLoS One. 2013 Sep 9;8(9):e73575. doi: 10.1371/journal.pone.0073575. eCollection 2013. PLoS One. 2013. PMID: 24039988 Free PMC article.
-
Heterologously expressed family 51 alpha-L-arabinofuranosidases from Oenococcus oeni and Lactobacillus brevis.Appl Environ Microbiol. 2011 Feb;77(4):1528-31. doi: 10.1128/AEM.01385-10. Epub 2010 Dec 17. Appl Environ Microbiol. 2011. PMID: 21169445 Free PMC article.
References
-
- Saha B. C. Alpha-L-arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol. Adv. 2000;18:403–423. - PubMed
-
- Sheehan J., Himmel M. Enzymes, energy, and the environment: a strategic perspective on the US Department of Energy's Research and Development Activities for Bioethanol. Biotechnol. Prog. 1999;15:817–827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

