The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin
- PMID: 15353600
- PMCID: PMC518786
- DOI: 10.1073/pnas.0405664101
The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin
Abstract
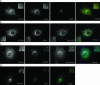
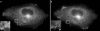
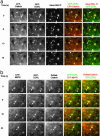
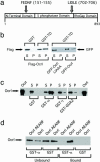
The subcellular localization of Ocrl, the inositol polyphosphate 5-phosphatase that is mutated in Lowe syndrome, was investigated by fluorescence microscopy. Ocrl was localized to endosomes and Golgi membranes along with clathrin, giantin, the mannose 6-phosphate receptor, transferrin, and the early endosomal antigen 1 endosomal marker in fixed cells. The endosomal localization of Ocrl was confirmed by live-cell time-lapse microscopy in which we monitored the dynamics of Ocrl on endosomes. GST binding assays show that Ocrl interacts with the clathrin terminal domain and the clathrin adaptor protein AP-2. Our findings suggest a role for Ocrl in endosomal receptor trafficking and sorting.
Figures

Similar articles
-
Rab35 GTPase Triggers Switch-like Recruitment of the Lowe Syndrome Lipid Phosphatase OCRL on Newborn Endosomes.Curr Biol. 2016 Jan 11;26(1):120-8. doi: 10.1016/j.cub.2015.11.040. Epub 2015 Dec 24. Curr Biol. 2016. PMID: 26725203
-
Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network.Mol Biol Cell. 2005 Aug;16(8):3467-79. doi: 10.1091/mbc.e05-02-0120. Epub 2005 May 25. Mol Biol Cell. 2005. PMID: 15917292 Free PMC article.
-
Clathrin terminal domain-ligand interactions regulate sorting of mannose 6-phosphate receptors mediated by AP-1 and GGA adaptors.J Biol Chem. 2014 Feb 21;289(8):4906-18. doi: 10.1074/jbc.M113.535211. Epub 2014 Jan 9. J Biol Chem. 2014. PMID: 24407285 Free PMC article.
-
Structure and function of the Lowe syndrome protein OCRL1.Traffic. 2005 Sep;6(9):711-9. doi: 10.1111/j.1600-0854.2005.00311.x. Traffic. 2005. PMID: 16101675 Review.
-
The role of the Lowe syndrome protein OCRL in the endocytic pathway.Biol Chem. 2015 Dec;396(12):1293-300. doi: 10.1515/hsz-2015-0180. Biol Chem. 2015. PMID: 26351914 Review.
Cited by
-
Phosphoinositides: tiny lipids with giant impact on cell regulation.Physiol Rev. 2013 Jul;93(3):1019-137. doi: 10.1152/physrev.00028.2012. Physiol Rev. 2013. PMID: 23899561 Free PMC article. Review.
-
A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway.Dev Cell. 2007 Sep;13(3):377-90. doi: 10.1016/j.devcel.2007.08.004. Dev Cell. 2007. PMID: 17765681 Free PMC article.
-
The PH domain proteins IPIP27A and B link OCRL1 to receptor recycling in the endocytic pathway.Mol Biol Cell. 2011 Mar 1;22(5):606-23. doi: 10.1091/mbc.E10-08-0730. Epub 2011 Jan 13. Mol Biol Cell. 2011. PMID: 21233288 Free PMC article.
-
Suppression of intestinal calcium entry channel TRPV6 by OCRL, a lipid phosphatase associated with Lowe syndrome and Dent disease.Am J Physiol Cell Physiol. 2012 May 15;302(10):C1479-91. doi: 10.1152/ajpcell.00277.2011. Epub 2012 Feb 29. Am J Physiol Cell Physiol. 2012. PMID: 22378746 Free PMC article.
-
Impaired neural development in a zebrafish model for Lowe syndrome.Hum Mol Genet. 2012 Apr 15;21(8):1744-59. doi: 10.1093/hmg/ddr608. Epub 2011 Dec 30. Hum Mol Genet. 2012. PMID: 22210625 Free PMC article.
References
-
- Attree, O., Olivos, I. M., Okabe, I., Bailey, L. C., Nelson, D. L., Lewis, R. A., McInnes, R. R. & Nussbaum, R. L. (1992) Nature 358, 239–242. - PubMed
-
- Nussbaum, R. (2000) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Sly, W.S., Childs, B., Beaudet, A. L., Valle, D., Kinzler, K. W. & Vogelstein, B. (McGraw–Hill, New York), 8th Ed., pp. 6257–6266.
-
- Majerus, P. W., Kisseleva, M. V. & Norris, F. A. (1999) J. Biol. Chem. 274, 10669–10672. - PubMed
-
- Zhang, X., Hartz, P. A., Philip, E., Racusen, L. C. & Majerus, P. W. (1998) J. Biol. Chem. 273, 1574–1582. - PubMed
-
- Suchy, S. F., Olivos-Glander, I. M. & Nussbaum, R. L. (1995) Hum. Mol. Genet. 4, 2245–2250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

