Aminoacylation properties of pathology-related human mitochondrial tRNA(Lys) variants
- PMID: 15100439
- PMCID: PMC1370574
- DOI: 10.1261/rna.5267604
Aminoacylation properties of pathology-related human mitochondrial tRNA(Lys) variants
Abstract
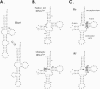
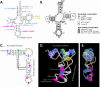
In vitro transcription has proven to be a successful tool for preparation of functional RNAs, especially in the tRNA field, in which, despite the absence of post-transcriptional modifications, transcripts are correctly folded and functionally active. Human mitochondrial (mt) tRNA(Lys) deviates from this principle and folds into various inactive conformations, due to the absence of the post-transcriptional modification m(1)A9 which hinders base-pairing with U64 in the native tRNA. Unavailability of a functional transcript is a serious drawback for structure/function investigations as well as in deciphering the molecular mechanisms by which point mutations in the mt tRNA(Lys) gene cause severe human disorders. Here, we show that an engineered in vitro transcribed "pseudo-WT" tRNA(Lys) variant is efficiently recognized by lysyl-tRNA synthetase and can substitute for the WT tRNA as a valuable reference molecule. This has been exploited in a systematic analysis of the effects on aminoacylation of nine pathology-related mutations described so far. The sole mutation located in a loop of the tRNA secondary structure, A8344G, does not affect aminoacylation efficiency. Out of eight mutations located in helical domains converting canonical Watson-Crick pairs into G-U pairs or C.A mismatches, six have no effect on aminoacylation (A8296G, U8316C, G8342A, U8356C, U8362G, G8363A), and two lead to drastic decreases (5000- to 7000-fold) in lysylation efficiencies (G8313A and G8328A). This screening, allowing for analysis of the primary impact level of all mutations affecting one tRNA under comparable conditions, indicates distinct molecular origins for different disorders.
Figures
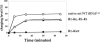



Similar articles
-
Comparative analysis of the pathogenic mechanisms associated with the G8363A and A8296G mutations in the mitochondrial tRNA(Lys) gene.Biochem J. 2005 May 1;387(Pt 3):773-8. doi: 10.1042/BJ20040949. Biochem J. 2005. PMID: 15554876 Free PMC article.
-
Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease.EMBO J. 2001 Sep 3;20(17):4794-802. doi: 10.1093/emboj/20.17.4794. EMBO J. 2001. PMID: 11532943 Free PMC article.
-
A double mutation (A8296G and G8363A) in the mitochondrial DNA tRNA (Lys) gene associated with myoclonus epilepsy with ragged-red fibers.Neurology. 1999 Jan 15;52(2):377-82. doi: 10.1212/wnl.52.2.377. Neurology. 1999. PMID: 9932960
-
Mitochondrial DNA G8363A mutation in the tRNA Lys gene: clinical, biochemical and pathological study.J Neurol Sci. 2009 Jun 15;281(1-2):85-92. doi: 10.1016/j.jns.2009.01.025. Epub 2009 Mar 10. J Neurol Sci. 2009. PMID: 19278689 Review.
-
Human mitochondrial diseases associated with tRNA wobble modification deficiency.RNA Biol. 2005 Apr;2(2):41-4. doi: 10.4161/rna.2.2.1610. Epub 2005 Apr 14. RNA Biol. 2005. PMID: 17132941 Review.
Cited by
-
Identity elements for the aminoacylation of metazoan mitochondrial tRNA(Arg) have been widely conserved throughout evolution and ensure the fidelity of the AGR codon reassignment.RNA Biol. 2014;11(10):1313-23. doi: 10.1080/15476286.2014.996094. RNA Biol. 2014. PMID: 25603118 Free PMC article.
-
Evidence for the presence of somatic mitochondrial DNA mutations in right atrial appendage tissues of coronary artery disease patients.Mol Genet Genomics. 2014 Aug;289(4):533-40. doi: 10.1007/s00438-014-0828-2. Epub 2014 Mar 7. Mol Genet Genomics. 2014. PMID: 24604425
-
Disease-associated mutations in mitochondrial precursor tRNAs affect binding, m1R9 methylation, and tRNA processing by mtRNase P.RNA. 2021 Apr;27(4):420-432. doi: 10.1261/rna.077198.120. Epub 2020 Dec 30. RNA. 2021. PMID: 33380464 Free PMC article.
-
Post-transcriptional nucleotide modification and alternative folding of RNA.Nucleic Acids Res. 2006 Feb 1;34(2):721-33. doi: 10.1093/nar/gkj471. Print 2006. Nucleic Acids Res. 2006. PMID: 16452298 Free PMC article. Review.
-
Mitochondrial tRNA 3' end metabolism and human disease.Nucleic Acids Res. 2004 Oct 11;32(18):5430-41. doi: 10.1093/nar/gkh884. Print 2004. Nucleic Acids Res. 2004. PMID: 15477393 Free PMC article. Review.
References
-
- Agris, P.F. 1996. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 53: 79–129. - PubMed
-
- Allen, J.F. and Raven, J.A. 1996. Free-radical-induced mutation vs redox regulation: Costs and benefits of genes in organelles. J. Mol. Evol. 42: 482–492. - PubMed
-
- Anderson, S., Bankier, A.T., Barrel, B.G., de Bruijn, M.H.L., Coulson, A.R., Drouin, J., Eperon, J.C., Nierlich, D.P., Roe, B.A., Sanger, F., et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. - PubMed
-
- Becker, H., Giegé, R., and Kern, D. 1996. Identity of prokaryotic and eukaryotic tRNAAsp for aminoacylation by aspartyl-tRNA synthetase from Thermus thermophilus. Biochemistry 35: 7447–7458. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
