New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control
- PMID: 15096624
- PMCID: PMC404060
- DOI: 10.1073/pnas.0308014101
New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control
Abstract
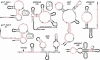
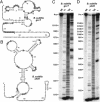
The expression of certain genes involved in fundamental metabolism is regulated by metabolite-binding "riboswitch" elements embedded within their corresponding mRNAs. We have identified at least six additional elements within the Bacillus subtilis genome that exhibit characteristics of riboswitch function (glmS, gcvT, ydaO/yuaA, ykkC/yxkD, ykoK, and yybP/ykoY). These motifs exhibit extensive sequence and secondary-structure conservation among many bacterial species and occur upstream of related genes. The element located upstream of the glmS gene in Gram-positive organisms functions as a metabolite-dependent ribozyme that responds to glucosamine-6-phosphate. Other motifs form complex folded structures when transcribed as RNA molecules and carry intrinsic terminator structures. These findings indicate that riboswitches serve as a major genetic regulatory mechanism for the control of metabolic genes in many microbial species.
Figures

Similar articles
-
Trans-acting glmS catalytic riboswitch: locked and loaded.RNA. 2007 Apr;13(4):468-77. doi: 10.1261/rna.341807. Epub 2007 Feb 5. RNA. 2007. PMID: 17283212 Free PMC article.
-
Challenges of ligand identification for riboswitch candidates.RNA Biol. 2011 Jan-Feb;8(1):5-10. doi: 10.4161/rna.8.1.13865. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21317561 Free PMC article.
-
Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions.RNA. 2006 Apr;12(4):607-19. doi: 10.1261/rna.2266506. Epub 2006 Feb 16. RNA. 2006. PMID: 16484375 Free PMC article.
-
[Riboswitches].Postepy Biochem. 2005;51(2):111-9. Postepy Biochem. 2005. PMID: 16209348 Review. Polish.
-
RNase P RNA of Mycoplasma capricolum.Mol Biol Rep. 1995-1996;22(2-3):125-9. doi: 10.1007/BF00988716. Mol Biol Rep. 1995. PMID: 8901498 Review.
Cited by
-
Comprehensive discovery of novel structured noncoding RNAs in 26 bacterial genomes.RNA Biol. 2021 Dec;18(12):2417-2432. doi: 10.1080/15476286.2021.1917891. Epub 2021 May 10. RNA Biol. 2021. PMID: 33970790 Free PMC article.
-
Do the P1 and P2 hairpins of the Guanidine-II riboswitch interact?Nucleic Acids Res. 2020 Oct 9;48(18):10518-10526. doi: 10.1093/nar/gkaa703. Nucleic Acids Res. 2020. PMID: 32857846 Free PMC article.
-
Imaging metabolite dynamics in living cells using a Spinach-based riboswitch.Proc Natl Acad Sci U S A. 2015 May 26;112(21):E2756-65. doi: 10.1073/pnas.1504354112. Epub 2015 May 11. Proc Natl Acad Sci U S A. 2015. PMID: 25964329 Free PMC article.
-
Nucleic Acid-Based Nanodevices in Biological Imaging.Annu Rev Biochem. 2016 Jun 2;85:349-73. doi: 10.1146/annurev-biochem-060815-014244. Annu Rev Biochem. 2016. PMID: 27294440 Free PMC article. Review.
-
Thermozymes: Synthetic RNA thermometers based on ribozyme activity.RNA Biol. 2013 Jun;10(6):1010-6. doi: 10.4161/rna.24482. Epub 2013 Apr 1. RNA Biol. 2013. PMID: 23595083 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

