ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating
- PMID: 15034580
- PMCID: PMC1995438
- DOI: 10.1038/ng1329
ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating
Abstract
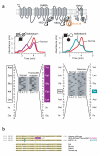
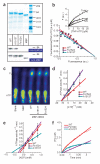
Stress tolerance of the heart requires high-fidelity metabolic sensing by ATP-sensitive potassium (K(ATP)) channels that adjust membrane potential-dependent functions to match cellular energetic demand. Scanning of genomic DNA from individuals with heart failure and rhythm disturbances due to idiopathic dilated cardiomyopathy identified two mutations in ABCC9, which encodes the regulatory SUR2A subunit of the cardiac K(ATP) channel. These missense and frameshift mutations mapped to evolutionarily conserved domains adjacent to the catalytic ATPase pocket within SUR2A. Mutant SUR2A proteins showed aberrant redistribution of conformations in the intrinsic ATP hydrolytic cycle, translating into abnormal K(ATP) channel phenotypes with compromised metabolic signal decoding. Defective catalysis-mediated pore regulation is thus a mechanism for channel dysfunction and susceptibility to dilated cardiomyopathy.
Figures


Similar articles
-
ATP-sensitive K+ channel channel/enzyme multimer: metabolic gating in the heart.J Mol Cell Cardiol. 2005 Jun;38(6):895-905. doi: 10.1016/j.yjmcc.2005.02.022. Epub 2005 Apr 14. J Mol Cell Cardiol. 2005. PMID: 15910874 Free PMC article. Review.
-
Cardiac KATP channels in health and disease.J Mol Cell Cardiol. 2005 Jun;38(6):937-43. doi: 10.1016/j.yjmcc.2005.02.026. Epub 2005 Apr 25. J Mol Cell Cardiol. 2005. PMID: 15910878 Free PMC article. Review.
-
Differential nucleotide regulation of KATP channels by SUR1 and SUR2A.J Mol Cell Cardiol. 2005 Sep;39(3):491-501. doi: 10.1016/j.yjmcc.2005.03.009. J Mol Cell Cardiol. 2005. PMID: 15893323
-
ATPase activity of the sulfonylurea receptor: a catalytic function for the KATP channel complex.FASEB J. 2000 Oct;14(13):1943-52. doi: 10.1096/fj.00-0027com. FASEB J. 2000. PMID: 11023978
-
Interaction of asymmetric ABCC9-encoded nucleotide binding domains determines KATP channel SUR2A catalytic activity.J Proteome Res. 2008 Apr;7(4):1721-8. doi: 10.1021/pr7007847. Epub 2008 Mar 1. J Proteome Res. 2008. PMID: 18311911 Free PMC article.
Cited by
-
Repression of the Central Splicing Regulator RBFox2 Is Functionally Linked to Pressure Overload-Induced Heart Failure.Cell Rep. 2015 Mar 10;10(9):1521-1533. doi: 10.1016/j.celrep.2015.02.013. Epub 2015 Mar 5. Cell Rep. 2015. PMID: 25753418 Free PMC article.
-
Determining the Likelihood of Disease Pathogenicity Among Incidentally Identified Genetic Variants in Rare Dilated Cardiomyopathy-Associated Genes.J Am Heart Assoc. 2022 Oct 4;11(19):e025257. doi: 10.1161/JAHA.122.025257. Epub 2022 Sep 21. J Am Heart Assoc. 2022. PMID: 36129056 Free PMC article.
-
KATP channel Kir6.2 E23K variant overrepresented in human heart failure is associated with impaired exercise stress response.Hum Genet. 2009 Dec;126(6):779-89. doi: 10.1007/s00439-009-0731-9. Hum Genet. 2009. PMID: 19685080 Free PMC article.
-
Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals.Genet Med. 2010 Nov;12(11):655-67. doi: 10.1097/GIM.0b013e3181f2481f. Genet Med. 2010. PMID: 20864896 Free PMC article.
-
Cardiac and vascular KATP channels in rats are activated by endogenous epoxyeicosatrienoic acids through different mechanisms.J Physiol. 2006 Sep 1;575(Pt 2):627-44. doi: 10.1113/jphysiol.2006.113985. Epub 2006 Jun 22. J Physiol. 2006. PMID: 16793897 Free PMC article.
References
-
- Inagaki N, et al. Reconstitution of IKATP: An inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270:1166–1170. - PubMed
-
- Inagaki N, et al. A family of sulfonylurea receptors determines the pharmacological properties of ATP-sensitive K+ channels. Neuron. 1996;16:1011–1017. - PubMed
-
- Bienengraeber M, et al. ATPase activity of the sulfonylurea receptor: A catalytic function for the KATP channel complex. FASEB J. 2000;14:1943–1952. - PubMed
-
- Matsuo M, Tanabe K, Kioka N, Amachi T, Ueda K. Different binding properties and affinities for ATP and ADP among sulfonylurea receptor subtypes, SUR1, SUR2A, and SUR2B. J. Biol. Chem. 2000;275:28757–28763. - PubMed
-
- Zingman LV, et al. Signaling in channel/enzyme multimers: ATPase transitions in SUR module gate ATP-sensitive K+ conductance. Neuron. 2001;31:233–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

