Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus
- PMID: 12851461
- PMCID: PMC166390
- DOI: 10.1073/pnas.1533026100
Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus
Abstract
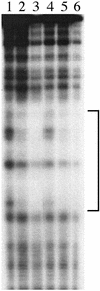
Myxococcus xanthus exhibits social behavior and multicellular development. FruA is an essential transcription factor for fruiting body development in M. xanthus. In the present study, the upstream promoter region was found to be necessary for the induction of fruA expression during development. A cis-acting element required for the induction was identified and was located between nucleotides -154 and -107 with respect to the transcription initiation site. In addition, it was found that two binding sites exist within this element of the fruA promoter. By using DNA affinity column chromatography containing the cis-acting element, a fruA promoter-binding protein was purified. The purified protein was shown by N-terminal sequence analysis to be identical to MrpC, a protein identified previously by transposon insertion mutagenesis as an essential locus for fruiting body development [Sun, H. & Shi, W. (2001) J. Bacteriol. 183, 4786-4795]. Furthermore, fruA mRNA was not detectable in the mrpC::km strain, demonstrating that MrpC is essential for fruA expression. Moreover, mutational analysis of the binding sites for MrpC in the fruA promoter indicates that binding of MrpC activates transcription of fruA in vivo. This report provides evidence for a direct molecular interaction involved in temporally regulated gene expression in M. xanthus.
Figures

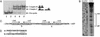



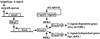
Similar articles
-
A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development.Mol Microbiol. 2006 Jun;60(5):1205-17. doi: 10.1111/j.1365-2958.2006.05178.x. Mol Microbiol. 2006. PMID: 16689796
-
Dual regulation with Ser/Thr kinase cascade and a His/Asp TCS in Myxococcus xanthus.Adv Exp Med Biol. 2008;631:111-21. doi: 10.1007/978-0-387-78885-2_7. Adv Exp Med Biol. 2008. PMID: 18792684 Review.
-
Transcription factor MrpC binds to promoter regions of hundreds of developmentally-regulated genes in Myxococcus xanthus.BMC Genomics. 2014 Dec 16;15:1123. doi: 10.1186/1471-2164-15-1123. BMC Genomics. 2014. PMID: 25515642 Free PMC article.
-
Regulation of FRUA expression during vegetative growth and development of Myxococcus xanthus.J Mol Microbiol Biotechnol. 2003;5(2):87-96. doi: 10.1159/000069979. J Mol Microbiol Biotechnol. 2003. PMID: 12736531
-
Intercellular signaling during fruiting-body development of Myxococcus xanthus.Annu Rev Microbiol. 1999;53:525-49. doi: 10.1146/annurev.micro.53.1.525. Annu Rev Microbiol. 1999. PMID: 10547700 Review.
Cited by
-
Development versus predation: Transcriptomic changes during the lifecycle of Myxococcus xanthus.Front Microbiol. 2022 Sep 26;13:1004476. doi: 10.3389/fmicb.2022.1004476. eCollection 2022. Front Microbiol. 2022. PMID: 36225384 Free PMC article.
-
Bacterial development in the fast lane.J Bacteriol. 2008 Jul;190(13):4373-6. doi: 10.1128/JB.00580-08. Epub 2008 May 9. J Bacteriol. 2008. PMID: 18469109 Free PMC article. No abstract available.
-
The dev Operon Regulates the Timing of Sporulation during Myxococcus xanthus Development.J Bacteriol. 2017 Apr 25;199(10):e00788-16. doi: 10.1128/JB.00788-16. Print 2017 May 15. J Bacteriol. 2017. PMID: 28264995 Free PMC article.
-
Mutation of self-binding sites in the promoter of the MrpC transcriptional regulator leads to asynchronous Myxococcus xanthus development.Front Microbiol. 2023 Nov 23;14:1293966. doi: 10.3389/fmicb.2023.1293966. eCollection 2023. Front Microbiol. 2023. PMID: 38075919 Free PMC article.
-
CRP-Like Transcriptional Regulator MrpC Curbs c-di-GMP and 3',3'-cGAMP Nucleotide Levels during Development in Myxococcus xanthus.mBio. 2021 Feb 22;13(1):e0004422. doi: 10.1128/mbio.00044-22. Epub 2022 Feb 15. mBio. 2021. PMID: 35164555 Free PMC article.
References
-
- Dworkin, M. & Kaiser, D. (1993) Myxobacteria II (Am. Soc. Microbiol., Washington, DC).
-
- Shimkets, L. J. (1999) Annu. Rev. Microbiol. 53, 525–549. - PubMed
-
- Horiuchi, T., Taoka, M., Isobe, T., Komano, T. & Inouye, S. (2002) J. Biol. Chem. 277, 26753–26760. - PubMed
-
- Ogawa, M., Fujitani, S., Mao, X., Inouye, S. & Komano, T. (1996) Mol. Microbiol. 22, 757–767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

