Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase
- PMID: 11593006
- PMCID: PMC59822
- DOI: 10.1073/pnas.211429998
Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase
Abstract
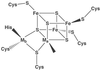
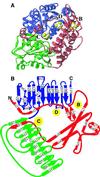
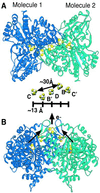
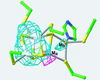
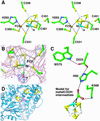
A crystal structure of the anaerobic Ni-Fe-S carbon monoxide dehydrogenase (CODH) from Rhodospirillum rubrum has been determined to 2.8-A resolution. The CODH family, for which the R. rubrum enzyme is the prototype, catalyzes the biological oxidation of CO at an unusual Ni-Fe-S cluster called the C-cluster. The Ni-Fe-S C-cluster contains a mononuclear site and a four-metal cubane. Surprisingly, anomalous dispersion data suggest that the mononuclear site contains Fe and not Ni, and the four-metal cubane has the form [NiFe(3)S(4)] and not [Fe(4)S(4)]. The mononuclear site and the four-metal cluster are bridged by means of Cys(531) and one of the sulfides of the cube. CODH is organized as a dimer with a previously unidentified [Fe(4)S(4)] cluster bridging the two subunits. Each monomer is comprised of three domains: a helical domain at the N terminus, an alpha/beta (Rossmann-like) domain in the middle, and an alpha/beta (Rossmann-like) domain at the C terminus. The helical domain contributes ligands to the bridging [Fe(4)S(4)] cluster and another [Fe(4)S(4)] cluster, the B-cluster, which is involved in electron transfer. The two Rossmann domains contribute ligands to the active site C-cluster. This x-ray structure provides insight into the mechanism of biological CO oxidation and has broader significance for the roles of Ni and Fe in biological systems.
Figures
Similar articles
-
Initial structure modification of tetrahedral to planar nickel(II) in a nickel-iron-sulfur cluster related to the C-cluster of carbon monoxide dehydrogenase.J Am Chem Soc. 2004 May 26;126(20):6448-59. doi: 10.1021/ja030627s. J Am Chem Soc. 2004. PMID: 15149242
-
Carbon monoxide induced decomposition of the active site [Ni-4Fe-5S] cluster of CO dehydrogenase.J Am Chem Soc. 2004 May 5;126(17):5382-7. doi: 10.1021/ja037776v. J Am Chem Soc. 2004. PMID: 15113209
-
New insights into the mechanism of nickel insertion into carbon monoxide dehydrogenase: analysis of Rhodospirillum rubrum carbon monoxide dehydrogenase variants with substituted ligands to the [Fe3S4] portion of the active-site C-cluster.J Biol Inorg Chem. 2005 Dec;10(8):903-12. doi: 10.1007/s00775-005-0043-z. Epub 2005 Nov 8. J Biol Inorg Chem. 2005. PMID: 16283394
-
The metalloclusters of carbon monoxide dehydrogenase/acetyl-CoA synthase: a story in pictures.J Biol Inorg Chem. 2004 Jul;9(5):511-5. doi: 10.1007/s00775-004-0563-y. Epub 2004 Jun 18. J Biol Inorg Chem. 2004. PMID: 15221484 Review.
-
A role for nickel-iron cofactors in biological carbon monoxide and carbon dioxide utilization.Curr Opin Chem Biol. 2011 Apr;15(2):276-83. doi: 10.1016/j.cbpa.2010.11.005. Epub 2010 Dec 2. Curr Opin Chem Biol. 2011. PMID: 21130022 Free PMC article. Review.
Cited by
-
Structural Insights into Microbial One-Carbon Metabolic Enzymes Ni-Fe-S-Dependent Carbon Monoxide Dehydrogenases and Acetyl-CoA Synthases.Biochemistry. 2022 Dec 20;61(24):2797-2805. doi: 10.1021/acs.biochem.2c00425. Epub 2022 Sep 22. Biochemistry. 2022. PMID: 36137563 Free PMC article. Review.
-
Life on the fringe: microbial adaptation to growth on carbon monoxide.F1000Res. 2018 Dec 27;7:F1000 Faculty Rev-1981. doi: 10.12688/f1000research.16059.1. eCollection 2018. F1000Res. 2018. PMID: 30647903 Free PMC article. Review.
-
Biome-specific distribution of Ni-containing carbon monoxide dehydrogenases.Extremophiles. 2022 Jan 20;26(1):9. doi: 10.1007/s00792-022-01259-y. Extremophiles. 2022. PMID: 35059858 Free PMC article.
-
Second and Outer Coordination Sphere Effects in Nitrogenase, Hydrogenase, Formate Dehydrogenase, and CO Dehydrogenase.Chem Rev. 2022 Jul 27;122(14):11900-11973. doi: 10.1021/acs.chemrev.1c00914. Epub 2022 Jul 18. Chem Rev. 2022. PMID: 35849738 Free PMC article. Review.
-
Comparative genomics in acid mine drainage biofilm communities reveals metabolic and structural differentiation of co-occurring archaea.BMC Genomics. 2013 Jul 17;14:485. doi: 10.1186/1471-2164-14-485. BMC Genomics. 2013. PMID: 23865623 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

