Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors
- PMID: 11585922
- PMCID: PMC99914
- DOI: 10.1128/MCB.21.21.7416-7428.2001
Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors
Abstract
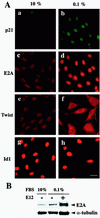
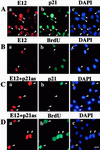
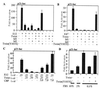
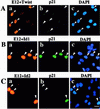
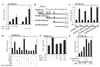
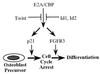
Cellular differentiation entails the coordination of cell cycle arrest and tissue-specific gene expression. We investigated the involvement of basic helix-loop-helix (bHLH) factors in differentiation of osteoblasts using the human osteoblastic cell line MG63. Serum starvation induced growth arrest at G1 phase, accompanied by expression of cyclin-dependent kinase inhibitor p21(WAF1/Cip1). Reporter assays with the p21 gene promoter demonstrated that the combination of E2A (E12 or E47) and coactivator CBP was responsible for p21 induction independent of p53. Twist inhibited E2A-CBP-dependent activation of the exogenous and endogenous p21 promoters. Ids similarly inhibited the exogenously transfected p21 promoter; however less antagonistic effect on the endogenous p21 promoter was observed. Twist was predominantly present in nuclei in MG63 cells growing in complete medium, while it localized mainly in the cytoplasm after serum starvation. The fibroblast growth factor receptor 3 gene (FGFR3), which generates signals leading to differentiation of osteoblasts, was found to be controlled by the same transcriptional regulation as the p21 gene. E2A and Twist influenced alkaline phosphatase expression, a consensus marker of osteoblast differentiation. Expression of E2A and FGFR3 was seen at the location of osteoblast differentiation in the calvaria of mouse embryos, implicating bHLH molecules in physiological osteoblast differentiation. These results demonstrate that a common regulatory system is involved in at least two distinct steps in osteoblastic differentiation. Our results also provide the molecular basis of Saethre-Chotzen syndrome, caused by mutations of the TWIST and FGFR3 genes.
Figures



Similar articles
-
Snail regulates p21(WAF/CIP1) expression in cooperation with E2A and Twist.Biochem Biophys Res Commun. 2004 Dec 24;325(4):1136-44. doi: 10.1016/j.bbrc.2004.10.148. Biochem Biophys Res Commun. 2004. PMID: 15555546
-
Basic helix-loop-helix proteins bind to TrkB and p21(Cip1) promoters linking differentiation and cell cycle arrest in neuroblastoma cells.Mol Cell Biol. 2004 Apr;24(7):2662-72. doi: 10.1128/MCB.24.7.2662-2672.2004. Mol Cell Biol. 2004. PMID: 15024057 Free PMC article.
-
Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription.J Biol Chem. 2003 Feb 28;278(9):7486-93. doi: 10.1074/jbc.M212248200. Epub 2002 Dec 19. J Biol Chem. 2003. PMID: 12493738
-
Effects of histone acetylation and DNA methylation on p21( WAF1) regulation.World J Gastroenterol. 2002 Jun;8(3):400-5. doi: 10.3748/wjg.v8.i3.400. World J Gastroenterol. 2002. PMID: 12046058 Free PMC article. Review.
-
Twist functions in mouse development.Int J Dev Biol. 2002;46(4):401-13. Int J Dev Biol. 2002. PMID: 12141426 Review.
Cited by
-
Upregulation of Twist-1 by NF-kappaB blocks cytotoxicity induced by chemotherapeutic drugs.Mol Cell Biol. 2007 Jun;27(11):3920-35. doi: 10.1128/MCB.01219-06. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403902 Free PMC article.
-
Molecular dissection of germline chromothripsis in a developmental context using patient-derived iPS cells.Genome Med. 2017 Jan 26;9(1):9. doi: 10.1186/s13073-017-0399-z. Genome Med. 2017. PMID: 28126037 Free PMC article.
-
Tbx1 regulates oral epithelial adhesion and palatal development.Hum Mol Genet. 2012 Jun 1;21(11):2524-37. doi: 10.1093/hmg/dds071. Epub 2012 Feb 27. Hum Mol Genet. 2012. PMID: 22371266 Free PMC article.
-
TRE17/ubiquitin-specific protease 6 (USP6) oncogene translocated in aneurysmal bone cyst blocks osteoblastic maturation via an autocrine mechanism involving bone morphogenetic protein dysregulation.J Biol Chem. 2010 Nov 19;285(47):37111-20. doi: 10.1074/jbc.M110.175133. Epub 2010 Sep 23. J Biol Chem. 2010. PMID: 20864534 Free PMC article.
-
Transcriptional regulation of the cyclin-dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells.Nucleic Acids Res. 2006;34(22):6472-87. doi: 10.1093/nar/gkl861. Epub 2006 Nov 27. Nucleic Acids Res. 2006. PMID: 17130157 Free PMC article.
References
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Bate M, Rushton E, Currie D A. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. - PubMed
-
- Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. - PubMed
-
- Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
