The nitrite reductase from Pseudomonas aeruginosa: essential role of two active-site histidines in the catalytic and structural properties
- PMID: 11226222
- PMCID: PMC30121
- DOI: 10.1073/pnas.041365298
The nitrite reductase from Pseudomonas aeruginosa: essential role of two active-site histidines in the catalytic and structural properties
Abstract
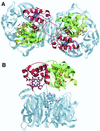
Cd(1) nitrite reductase catalyzes the conversion of nitrite to NO in denitrifying bacteria. Reduction of the substrate occurs at the d(1)-heme site, which faces on the distal side some residues thought to be essential for substrate binding and catalysis. We report the results obtained by mutating to Ala the two invariant active site histidines, His-327 and His-369, of the enzyme from Pseudomonas aeruginosa. Both mutants have lost nitrite reductase activity but maintain the ability to reduce O(2) to water. Nitrite reductase activity is impaired because of the accumulation of a catalytically inactive form, possibly because the productive displacement of NO from the ferric d(1)-heme iron is impaired. Moreover, the two distal His play different roles in catalysis; His-369 is absolutely essential for the stability of the Michaelis complex. The structures of both mutants show (i) the new side chain in the active site, (ii) a loss of density of Tyr-10, which slipped away with the N-terminal arm, and (iii) a large topological change in the whole c-heme domain, which is displaced 20 A from the position occupied in the wild-type enzyme. We conclude that the two invariant His play a crucial role in the activity and the structural organization of cd(1) nitrite reductase from P. aeruginosa.
Figures


Similar articles
-
Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.J Mol Biol. 2001 Sep 21;312(3):541-54. doi: 10.1006/jmbi.2001.4986. J Mol Biol. 2001. PMID: 11563915
-
Dynamic hydrogen-bonding network in the distal pocket of the nitrosyl complex of Pseudomonas aeruginosa cd1 nitrite reductase.J Am Chem Soc. 2011 Mar 9;133(9):3043-55. doi: 10.1021/ja109688w. Epub 2011 Feb 10. J Am Chem Soc. 2011. PMID: 21309511
-
Nitrate reductase of Neurospora crassa: the functional role of individual amino acids in the heme domain as examined by site-directed mutagenesis.Mol Gen Genet. 1993 Aug;240(2):221-30. doi: 10.1007/BF00277060. Mol Gen Genet. 1993. PMID: 8355655
-
NO production by Pseudomonas aeruginosa cd1 nitrite reductase.IUBMB Life. 2003 Oct-Nov;55(10-11):617-21. doi: 10.1080/15216540310001628672. IUBMB Life. 2003. PMID: 14711008 Review.
-
New insights into the activity of Pseudomonas aeruginosa cd1 nitrite reductase.Biochem Soc Trans. 2008 Dec;36(Pt 6):1155-9. doi: 10.1042/BST0361155. Biochem Soc Trans. 2008. PMID: 19021515 Review.
Cited by
-
Nitrite and nitrate chemical biology and signalling.Br J Pharmacol. 2019 Jan;176(2):228-245. doi: 10.1111/bph.14484. Epub 2018 Oct 3. Br J Pharmacol. 2019. PMID: 30152056 Free PMC article. Review.
-
Rational design of a nitrite reductase based on myoglobin: a molecular modeling and dynamics simulation study.J Mol Model. 2012 Sep;18(9):4409-15. doi: 10.1007/s00894-012-1451-y. Epub 2012 May 16. J Mol Model. 2012. PMID: 22588586
-
Higher diversity and abundance of denitrifying microorganisms in environments than considered previously.ISME J. 2015 Sep;9(9):1954-65. doi: 10.1038/ismej.2015.9. Epub 2015 Mar 10. ISME J. 2015. PMID: 25756678 Free PMC article.
-
Phylogenetics and environmental distribution of nitric oxide-forming nitrite reductases reveal their distinct functional and ecological roles.ISME Commun. 2024 Feb 2;4(1):ycae020. doi: 10.1093/ismeco/ycae020. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38584645 Free PMC article.
-
The functional role of the structure of the dioxo-isobacteriochlorin in the catalytic site of cytochrome cd1 for the reduction of nitrite.Chem Sci. 2016 Apr 1;7(4):2896-2906. doi: 10.1039/c5sc04825g. Epub 2016 Jan 20. Chem Sci. 2016. PMID: 30090283 Free PMC article.
References
-
- Averill B A. Chem Rev. 1996;96:2951–2964. - PubMed
-
- Cutruzzolà F. Biochim Biophys Acta. 1999;4730:1–19. - PubMed
-
- Nurizzo D, Silvestrini M C, Mathieu M, Cutruzzolà F, Bourgeois D, Fülop V, Hajdu J, Brunori M, Tegoni M, Cambillau C. Structure (London) 1997;5:1157–1171. - PubMed
-
- Nurizzo D, Cutruzzolà F, Arese M, Bourgeois D, Brunori M, Tegoni M, Cambillau C. Biochemistry. 1998;37:13987–13996. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

