The COG database: new developments in phylogenetic classification of proteins from complete genomes
- PMID: 11125040
- PMCID: PMC29819
- DOI: 10.1093/nar/29.1.22
The COG database: new developments in phylogenetic classification of proteins from complete genomes
Abstract
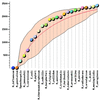
The database of Clusters of Orthologous Groups of proteins (COGs), which represents an attempt on a phylogenetic classification of the proteins encoded in complete genomes, currently consists of 2791 COGs including 45 350 proteins from 30 genomes of bacteria, archaea and the yeast Saccharomyces cerevisiae (http://www.ncbi.nlm.nih. gov/COG). In addition, a supplement to the COGs is available, in which proteins encoded in the genomes of two multicellular eukaryotes, the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster, and shared with bacteria and/or archaea were included. The new features added to the COG database include information pages with structural and functional details on each COG and literature references, improvements of the COGNITOR program that is used to fit new proteins into the COGs, and classification of genomes and COGs constructed by using principal component analysis.
Figures

Similar articles
-
The COG database: a tool for genome-scale analysis of protein functions and evolution.Nucleic Acids Res. 2000 Jan 1;28(1):33-6. doi: 10.1093/nar/28.1.33. Nucleic Acids Res. 2000. PMID: 10592175 Free PMC article.
-
The COG database: an updated version includes eukaryotes.BMC Bioinformatics. 2003 Sep 11;4:41. doi: 10.1186/1471-2105-4-41. Epub 2003 Sep 11. BMC Bioinformatics. 2003. PMID: 12969510 Free PMC article.
-
COG database update: focus on microbial diversity, model organisms, and widespread pathogens.Nucleic Acids Res. 2021 Jan 8;49(D1):D274-D281. doi: 10.1093/nar/gkaa1018. Nucleic Acids Res. 2021. PMID: 33167031 Free PMC article.
-
Functional genomics and enzyme evolution. Homologous and analogous enzymes encoded in microbial genomes.Genetica. 1999;106(1-2):159-70. doi: 10.1023/a:1003705601428. Genetica. 1999. PMID: 10710722 Review.
-
A genomic perspective on protein families.Science. 1997 Oct 24;278(5338):631-7. doi: 10.1126/science.278.5338.631. Science. 1997. PMID: 9381173 Review.
Cited by
-
The microbiome of the Black Sea water column analyzed by shotgun and genome centric metagenomics.Environ Microbiome. 2021 Mar 16;16(1):5. doi: 10.1186/s40793-021-00374-1. Environ Microbiome. 2021. PMID: 33902743 Free PMC article.
-
De Novo Assembly and Transcriptome Analysis of Bulb Onion (Allium cepa L.) during Cold Acclimation Using Contrasting Genotypes.PLoS One. 2016 Sep 14;11(9):e0161987. doi: 10.1371/journal.pone.0161987. eCollection 2016. PLoS One. 2016. PMID: 27627679 Free PMC article.
-
The draft genome of the Temminck's tragopan (Tragopan temminckii) with evolutionary implications.BMC Genomics. 2023 Dec 7;24(1):751. doi: 10.1186/s12864-023-09857-6. BMC Genomics. 2023. PMID: 38062370 Free PMC article.
-
Genome-wide transcriptional responses of two metal-tolerant symbiotic Mesorhizobium isolates to zinc and cadmium exposure.BMC Genomics. 2013 Apr 30;14:292. doi: 10.1186/1471-2164-14-292. BMC Genomics. 2013. PMID: 23631387 Free PMC article.
-
Draft genome sequence of an ammonia-oxidizing archaeon, "Candidatus Nitrosopumilus koreensis" AR1, from marine sediment.J Bacteriol. 2012 Dec;194(24):6940-1. doi: 10.1128/JB.01857-12. J Bacteriol. 2012. PMID: 23209206 Free PMC article.
References
-
- Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. - PubMed
-
- Fitch W.M. (1970) Distinguishing homologous from analogous proteins. Syst. Zool., 19, 99–106. - PubMed
-
- Kawarabayasi Y., Hino,Y., Horikawa,H., Yamazaki,S., Haikawa,Y., Jin-no,K., Takahashi,M., Sekine,M., Baba,S., Ankai,A. et al. (1999) Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res., 6, 83–101. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

