The genomic region encompassing the nephropathic cystinosis gene (CTNS): complete sequencing of a 200-kb segment and discovery of a novel gene within the common cystinosis-causing deletion
- PMID: 10673275
- PMCID: PMC310836
- DOI: 10.1101/gr.10.2.165
The genomic region encompassing the nephropathic cystinosis gene (CTNS): complete sequencing of a 200-kb segment and discovery of a novel gene within the common cystinosis-causing deletion
Abstract
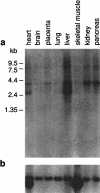
Nephropathic cystinosis is an autosomal recessive disorder caused by the defective transport of cystine out of lysosomes. Recently, the causative gene (CTNS) was identified and presumed to encode an integral membrane protein called cystinosin. Many of the disease-associated mutations in CTNS are deletions, including one >55 kb in size that represents the most common cystinosis allele encountered to date. In an effort to determine the precise genomic organization of CTNS and to gain sequence-based insight about the DNA within and flanking cystinosis-associated deletions, we mapped and sequenced the region of human chromosome 17p13 encompassing CTNS. Specifically, a bacterial artificial chromosome (BAC)-based physical map spanning CTNS was constructed by sequence-tagged site (STS)-content mapping. The resulting BAC contig provided the relative order of 43 STSs. Two overlapping BACs, which together contain all of the CTNS exons as well as extensive amounts of flanking DNA, were selected and subjected to shotgun sequencing. A total of 200,237 bp of contiguous, high-accuracy sequence was generated. Analysis of the resulting data revealed a number of interesting features about this genomic region, including the long-range organization of CTNS, insight about the breakpoints and intervening DNA associated with the common cystinosis-causing deletion, and structural information about five genes neighboring CTNS (human ortholog of rat vanilloid receptor subtype 1 gene, CARKL, TIP-1, P2X5, and HUMINAE). In particular, sequence analysis detected the presence of a novel gene (CARKL) residing within the most common cystinosis-causing deletion. This gene encodes a previously unknown protein that is predicted to function as a carbohydrate kinase. Interestingly, both CTNS and CARKL are absent in nearly half of all cystinosis patients (i.e., those homozygous for the common deletion). [The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF168787 and AF163573.]
Figures

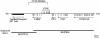
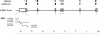
Similar articles
-
The promoter of a lysosomal membrane transporter gene, CTNS, binds Sp-1, shares sequences with the promoter of an adjacent gene, CARKL, and causes cystinosis if mutated in a critical region.Am J Hum Genet. 2001 Oct;69(4):712-21. doi: 10.1086/323484. Epub 2001 Aug 14. Am J Hum Genet. 2001. PMID: 11505338 Free PMC article.
-
A novel gene encoding an integral membrane protein is mutated in nephropathic cystinosis.Nat Genet. 1998 Apr;18(4):319-24. doi: 10.1038/ng0498-319. Nat Genet. 1998. PMID: 9537412
-
Sedoheptulokinase deficiency due to a 57-kb deletion in cystinosis patients causes urinary accumulation of sedoheptulose: elucidation of the CARKL gene.Hum Mutat. 2008 Apr;29(4):532-6. doi: 10.1002/humu.20685. Hum Mutat. 2008. PMID: 18186520
-
CTNS mutations in patients with cystinosis.Hum Mutat. 1999;14(6):454-8. doi: 10.1002/(SICI)1098-1004(199912)14:6<454::AID-HUMU2>3.0.CO;2-H. Hum Mutat. 1999. PMID: 10571941 Review.
-
[From gene to disease: cystinosis].Ned Tijdschr Geneeskd. 2004 Mar 6;148(10):476-8. Ned Tijdschr Geneeskd. 2004. PMID: 15042893 Review. Dutch.
Cited by
-
Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits.Nat Genet. 2021 Jun;53(6):779-786. doi: 10.1038/s41588-021-00865-4. Epub 2021 May 10. Nat Genet. 2021. PMID: 33972781
-
Cognition in nephropathic cystinosis: pattern of expression in heterozygous carriers.Am J Med Genet A. 2012 Aug;158A(8):1902-8. doi: 10.1002/ajmg.a.35467. Epub 2012 Jul 11. Am J Med Genet A. 2012. PMID: 22786804 Free PMC article.
-
Late-onset nephropathic cystinosis: clinical presentation, outcome, and genotyping.Clin J Am Soc Nephrol. 2008 Jan;3(1):27-35. doi: 10.2215/CJN.01740407. Clin J Am Soc Nephrol. 2008. PMID: 18178779 Free PMC article.
-
Synthesis of 4-Deoxy-4-Fluoro-d-Sedoheptulose: A Promising New Sugar to Apply the Principle of Metabolic Trapping.Chemistry. 2023 Nov 8;29(62):e202302277. doi: 10.1002/chem.202302277. Epub 2023 Sep 28. Chemistry. 2023. PMID: 37552007 Free PMC article.
-
New aspects of the pathogenesis of cystinosis.Pediatr Nephrol. 2003 Mar;18(3):207-15. doi: 10.1007/s00467-003-1077-5. Epub 2003 Feb 27. Pediatr Nephrol. 2003. PMID: 12644911 Review.
References
-
- Anikster Y, Lucero C, Touchman JW, Huizing M, McDowell G, Shotelersuk V, Green ED, Gahl WA. Identification and detection of the common 65-kb deletion breakpoint in the nephropathic cystinosis gene (CTNS) Mol Genet Metab. 1999;66:111–116. - PubMed
-
- Birren B, Mancino V, Shizuya H. Bacterial artificial chromosomes. In: Birren B, et al., editors. Genome Analysis Vol. 3, Cloning systems. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 241–295.
-
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. - PubMed
-
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
