Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava
- PMID: 10482497
- PMCID: PMC94076
- DOI: 10.1128/JB.181.18.5581-5590.1999
Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava
Abstract
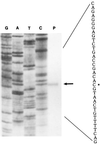
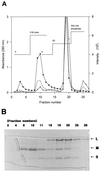
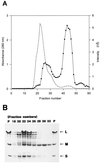
Carbon monoxide dehydrogenases (CO-DH) are the enzymes responsible for the oxidation of CO to carbon dioxide in carboxydobacteria and consist of three nonidentical subunits containing molybdopterin flavin adenine dinucleotide (FAD), and two different iron-sulfur clusters (O. Meyer, K. Frunzke, D. Gadkari, S. Jacobitz, I. Hugendieck, and M. Kraut, FEMS Microbiol. Rev. 87:253-260, 1990). The three structural genes of CO-DH in Hydrogenophaga pseudoflava were cloned and characterized. The genes were clustered on the chromosome in the transcriptional order cutM-cutS-cutL. The cloned cutM, cutS, and cutL genes had open reading frames of 864, 492, and 2,412 nucleotides, coding for proteins with calculated molecular weights of 30,694, 17,752, and 87,224, respectively. The overall identities in the nucleotide sequence of the genes and the amino acid sequence of the subunits with those of other carboxydobacteria were 64.5 to 74.3% and 62.8 to 72.3%, respectively. Primer extension analysis revealed that the transcriptional start site of the genes was the nucleotide G located 47 bp upstream of the cutM start codon. The deduced amino acid sequences of the three subunits of CO-DH implied the presence of molybdenum cofactor, FAD, and iron-sulfur centers in CutL, CutM, and CutS, respectively. Fluorometric analysis coupled with denaturing polyacrylamide gel electrophoresis of fractions from hydroxyapatite column chromatography in the presence of 8 M urea of active CO-DH and from gel filtration of spontaneously inactivated enzyme revealed that the large and medium subunits of CO-DH in H. pseudoflava bind molybdopterin and FAD cofactors, respectively. Iron-sulfur centers of the enzyme were identified to be present in the small subunit on the basis of the iron content in each subunit eluted from the denaturing polyacrylamide gels.
Figures




Similar articles
-
The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.J Mol Biol. 2000 Sep 1;301(5):1221-35. doi: 10.1006/jmbi.2000.4023. J Mol Biol. 2000. PMID: 10966817
-
Molecular characterization of the gene cluster coxMSL encoding the molybdenum-containing carbon monoxide dehydrogenase of Oligotropha carboxidovorans.J Bacteriol. 1995 Apr;177(8):2197-203. doi: 10.1128/jb.177.8.2197-2203.1995. J Bacteriol. 1995. PMID: 7721710 Free PMC article.
-
The tungsten formylmethanofuran dehydrogenase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic for enzymes containing molybdopterin dinucleotide.Eur J Biochem. 1995 Dec 15;234(3):910-20. doi: 10.1111/j.1432-1033.1995.910_a.x. Eur J Biochem. 1995. PMID: 8575452
-
The role of Se, Mo and Fe in the structure and function of carbon monoxide dehydrogenase.Biol Chem. 2000 Sep-Oct;381(9-10):865-76. doi: 10.1515/BC.2000.108. Biol Chem. 2000. PMID: 11076018 Review.
-
Molybdenum-cofactor-containing enzymes: structure and mechanism.Annu Rev Biochem. 1997;66:233-67. doi: 10.1146/annurev.biochem.66.1.233. Annu Rev Biochem. 1997. PMID: 9242907 Review.
Cited by
-
Physiological, ecological, and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus.Appl Environ Microbiol. 2007 Feb;73(4):1266-76. doi: 10.1128/AEM.01724-06. Epub 2006 Dec 1. Appl Environ Microbiol. 2007. PMID: 17142374 Free PMC article.
-
Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity.Appl Environ Microbiol. 2003 Dec;69(12):7257-65. doi: 10.1128/AEM.69.12.7257-7265.2003. Appl Environ Microbiol. 2003. PMID: 14660374 Free PMC article.
-
CO dehydrogenase genes found in metagenomic fosmid clones from the deep mediterranean sea.Appl Environ Microbiol. 2009 Dec;75(23):7436-44. doi: 10.1128/AEM.01283-09. Epub 2009 Oct 2. Appl Environ Microbiol. 2009. PMID: 19801465 Free PMC article.
-
The mononuclear molybdenum enzymes.Chem Rev. 2014 Apr 9;114(7):3963-4038. doi: 10.1021/cr400443z. Epub 2014 Jan 28. Chem Rev. 2014. PMID: 24467397 Free PMC article. Review. No abstract available.
-
The aerobic CO dehydrogenase from Oligotropha carboxidovorans.J Biol Inorg Chem. 2015 Mar;20(2):243-51. doi: 10.1007/s00775-014-1188-4. Epub 2014 Aug 26. J Biol Inorg Chem. 2015. PMID: 25156151 Review.
References
-
- Amaya Y, Yamazaki K, Sato M, Noda K, Nishino T, Nishino T. Proteolytic conversion of xanthine dehydrogenase from the NAD-dependent type to the O2-dependent type. J Biol Chem. 1990;265:14170–14175. - PubMed
-
- Biotech Laboratories Inc. Biotech Laboratories bulletin no. 27. Houston, Tex: Biotech Laboratories Inc.; 1992.
-
- Black G W, Lyons C M, Williams E, Colby J, Kehoe M, O’Reilly C. Cloning and expression of the carbon monoxide dehydrogenase genes from Pseudomonas thermocarboxydovorans strain C2. FEMS Microbiol Lett. 1990;70:249–254. - PubMed
-
- Blase M, Bruntner C, Tshisuaka B, Fetzner S, Lingens F. Cloning, expression, and sequence analysis of the three genes encoding quinoline 2-oxidoreductase from Pseudomonas putida 86. J Biol Chem. 1996;271:23068–23079. - PubMed
-
- Boehringer-Mannheim. The Dig system user’s guide. Indianapolis, Ind: Boehringer-Mannheim; 1993.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

